Principal Investigator

Staff
Research topic
Chromatin structure and epigenetic mechanisms for cancer growth and resistance to therapy.
Background
Recent advances in sequencing technologies and gene editing have revealed that cancer aggressiveness and resistance to standard treatments are strongly influenced by epigenetic factors, which regulate gene expression beyond the DNA sequence itself.
Among epigenetic factors, the three-dimensional structure of the chromatin plays a prominent role. Chromatin conformation is a strictly regulated process that governs accessibility to the underlying DNA sequence and thereby guides cellular processes critical for maintaining cellular identity and function. Other than regulating the expression of oncogenes and tumor suppressors, chromatin architecture is instructing many cancer-associated phenotypes, such as metastasis, therapy resistance, and immune evasion.
Research achievements
The projects in our lab aim to understand the epigenetic mechanisms that determine cancer behavior, with a particular focus on chromatin alterations during cancer development and resistance to therapy.
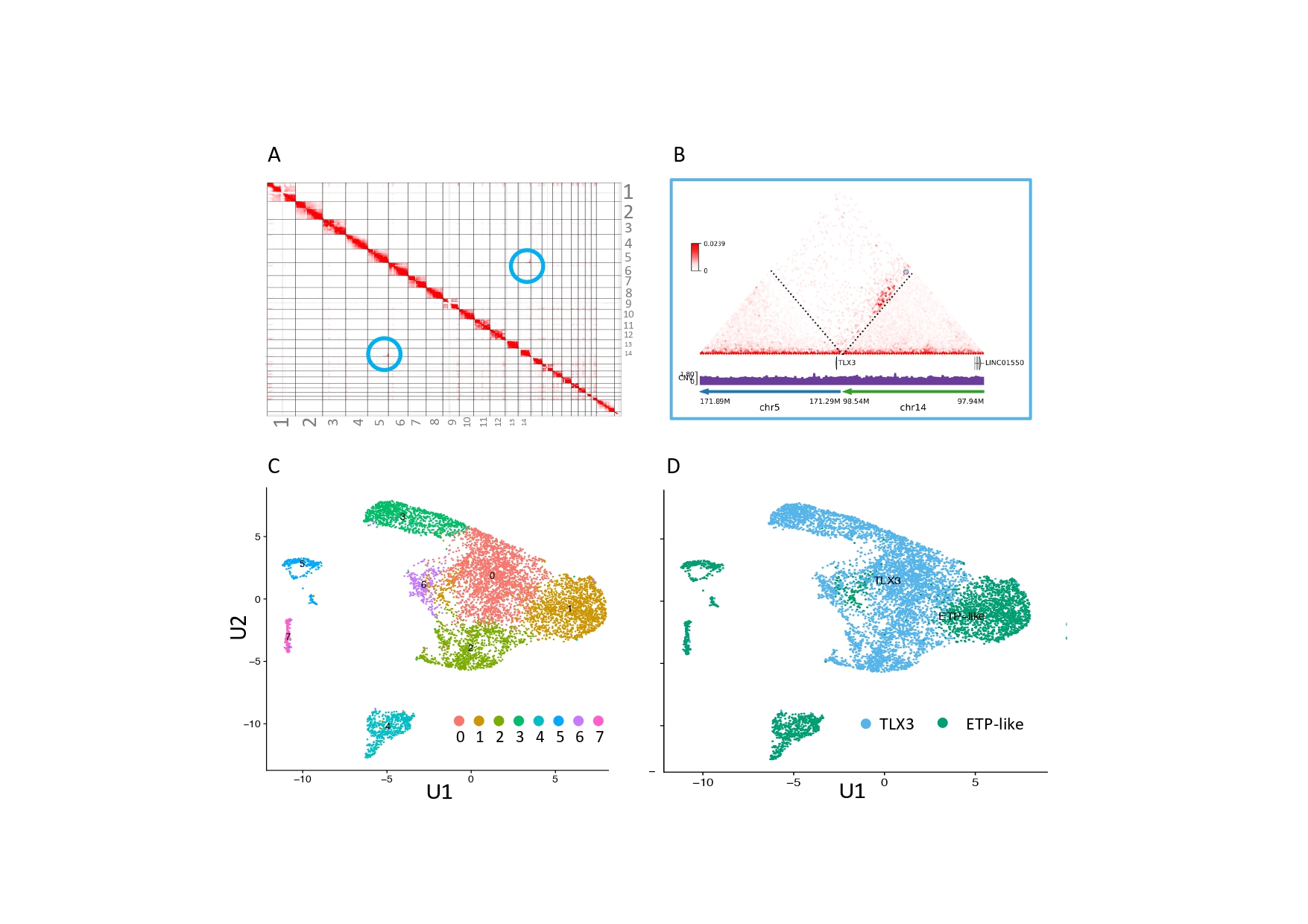
We have initially analyzed these aspects in the context of pediatric acute T lymphoblastic leukemia (T-ALL), where we demonstrated that chromatin architecture is subverted when a normal T cell progenitor is transformed into a leukemic blast. This is often accompanied by the disruption of boundary elements that favor the “rewiring” of the normal promoter-enhancer interactions, thus causing the promoters of proto-oncogenes to become activated by distal enhancers. A genome-wide characterization of chromatin architecture rearrangements across several patient groups has shown that specific conformations correspond to more aggressive subtypes, which in turn associate with increased resistance to therapy. We are now aiming at expanding these observations beyond pediatric leukemia, since we believe that similar mechanisms will apply to virtually every type of cancer.
Conclusions and perspectives
Our experimental approaches integrate genome-wide epigenetic screenings and advanced computational algorithms with the goal of better understanding the causes of cellular transformation and drug resistance. The findings of our studies will reveal novel biomarkers and regulators of resistance, thus potentially offering new targets for therapy and improving clinical outcomes, especially in subsets of patients that are challenging to treat.
Publications
At this link, you can find all the scientific publications of the Principal Investigator.
Selected publications
Gambi G*, Boccalatte F*@, Rodriguez-Hernaez J*, Lin Z, Nadorp B, Polyzos A, Tan J, Avrampou K, Inghirami G, Kentsis A, Apostolou E, Aifantis I, Tsirigos A@
Wang Q*, Boccalatte F*, Xu J, Gambi G, Nadorp B, Akter F, Mullin C, Melnick AF, Choe E, McCarter AC, Jerome NA, Chen S, Lin K, Khan S, Kodgule R, Sussman JH, Pölönen P, Rodriguez-Hernaez J, Narang S, Avrampou K, King B, Tsirigos A, Ryan RJH, Mullighan CG, Teachey DT, Tan K, Aifantis I, Chiang MY.
Parlani M*, Boccalatte F* @ , Yeaton A, Wang F, Zhang J, Aifantis I, Dondossola E# @
King B*, Boccalatte F*, Moran-Crusio K, Wolf E, Wang J, Kayembe C, Lazaris C, Yu X, Aranda-Orgilles B, Lasorella A and Aifantis I.
Nucera S*, Giustacchini A*, Boccalatte F*, Calabria A, Fanciullo C, Plati T, Ranghetti A, Garcia-Manteiga JM, Cittaro D, Benedicenti F, Lechman ER, Dick JE, Ponzoni M, Ciceri F, Montini E, Gentner B and Naldini L.